library(readxl)
library(tidyverse)
library(ggplot2)
fung <- read_excel("fung_myc.xlsx")
library(DT)
datatable(fung)Data analysis
The analysis of data from the in vitro experiment was conducted as follows:
#exploratory analysis
fung2 <- fung |>
group_by(fungicide, dose, time) |>
summarise(growth_mean = mean(growth))
pfung2 <- fung2 |>
ggplot(aes(dose, growth_mean, group = time,
color = factor(time)))+
geom_point()+
geom_line()+
facet_wrap(~fungicide) +
scale_y_continuous(n.breaks = 5, limits = c(0,5)) +
labs(x = "Dose (μg a.i. mL)", y = "Mycelial growth (cm)", title= "Effect of fungicides on mycelial growth", color = "Experiment" ) +
theme_minimal()+
theme(plot.title = element_text(hjust = 0.5))
pfung2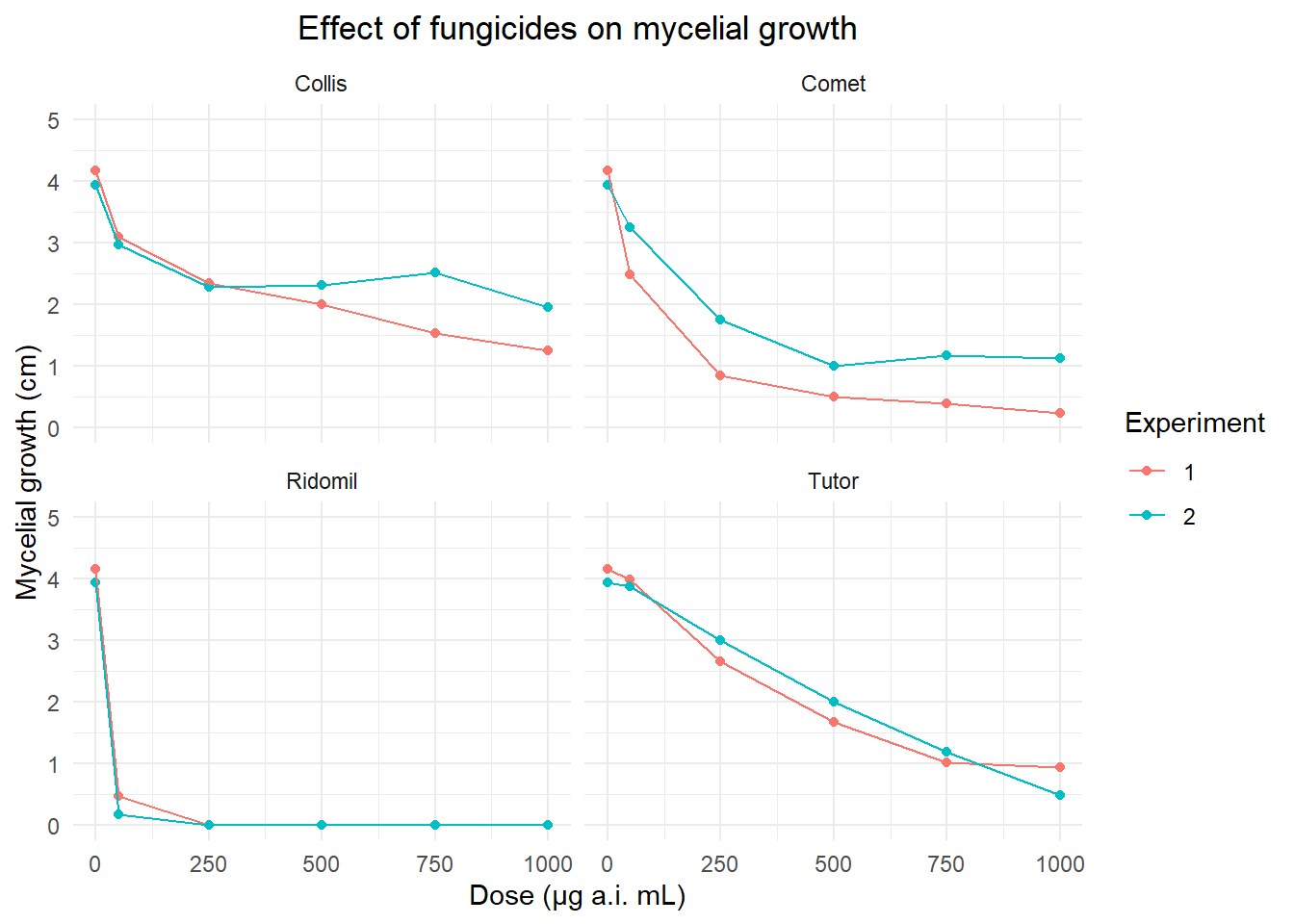
library(ggplot2)
library(dplyr)
library(patchwork)
fung2 <- fung |>
group_by(fungicide, dose, time) |>
summarise(growth_mean = mean(growth))
# Criaremos um gráfico separado para cada fungicida usando purrr::map e salvaremos os gráficos em uma lista
library(purrr)
# Vetor com os rótulos para cada fungicida
anotations <- c("EC50 = 67.26", "EC50 = 90.47", "EC50= 22 ", "EC50= 427.91")
plot_list <- fung2 %>%
# Converter a coluna "fungicide" para fator e recodificar os nomes
mutate(fungicide = factor(fungicide,
levels = c("Ridomil", "Comet", "Tutor", "Collis"),
labels = c("metalaxyl+mancozeb", "pyraclostrobin", "copper hydroxide", "boscalide+cresoxim-methyl"))) %>%
group_by(fungicide) %>%
group_split() %>%
map2(anotations, ~{
p <- ggplot(data = .x, aes(dose, growth_mean, group = time, color = factor(time))) +
geom_point() +
geom_line() +
scale_y_continuous(n.breaks = 5, limits = c(0, 5)) +
labs(x = "Dose (μg a.i. mL)", y = "Mycelial growth (cm)",
title = paste(unique(.x$fungicide)),
color = "Experiment") +
theme_minimal() +
theme(
plot.title = element_text(size = 10, hjust = 0.5),
axis.text.x = element_text(size = 7),
axis.text.y = element_text(size = 7),
axis.title.x = element_text(size = 7),
axis.title.y = element_text(size = 7),
legend.title = element_text(size = 7),
legend.text = element_text(size = 7),
plot.subtitle = element_text(size = 7),
plot.caption = element_text(size = 5)
)
p <- p + annotate("text", x = Inf, y = Inf, hjust = 1, vjust = 1,
label = .y, size = 2.5, color = "black")
return(p)
})
# Use o patchwork para unir os gráficos
final_plot <- wrap_plots(plotlist = plot_list)
# Imprimir o gráfico final
print(final_plot)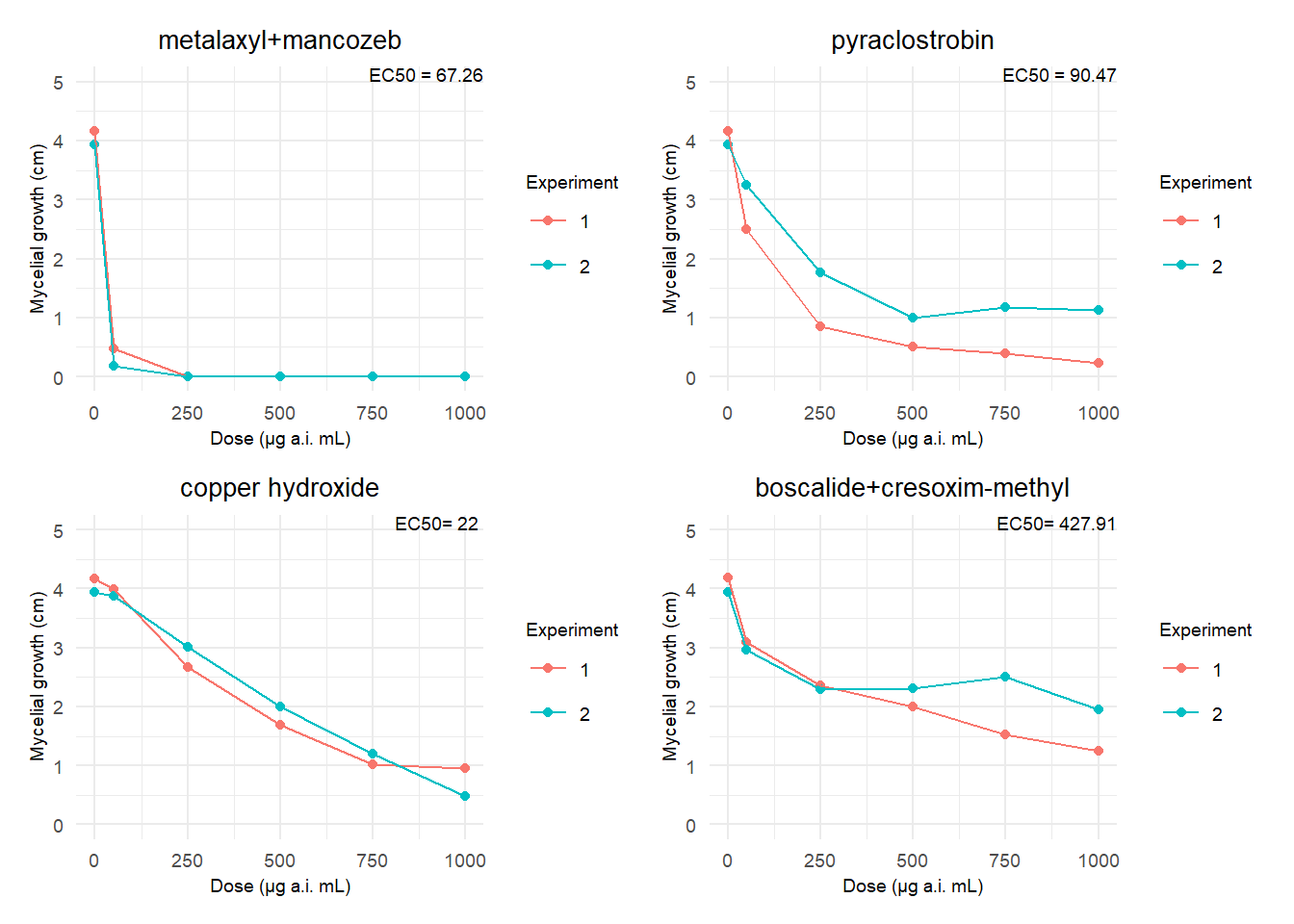
ggsave("finalplot_vitro.png",
bg = "white", width = 6, height= 4)library(ggplot2)
library(dplyr)
library(patchwork)
# Seu código para calcular o fung2 permanece o mesmo
fung2 <- fung |>
group_by(fungicide, dose, time) |>
summarise(growth_mean = mean(growth))
# Criaremos um gráfico separado para cada fungicida usando purrr::map e salvaremos os gráficos em uma lista
library(purrr)
# Vetor com os rótulos para cada fungicida
anotations <- c("EC50 = 67.26", "EC50 = 90.47", "EC50= 22 ", "EC50= 427.91")
plot_list <- fung2 %>%
group_by(fungicide) %>%
group_split() %>% # Corrigindo o uso do group_split
map2(anotations, ~{
# Criar o gráfico individual para cada fungicida
p <- ggplot(data = .x, aes(dose, growth_mean, group = time, color = factor(time))) +
geom_point() +
geom_line() +
scale_y_continuous(n.breaks = 5, limits = c(0, 5)) +
labs(x = "Dose (μg a.i. mL)", y = "Mycelial growth (cm)",
title = paste(unique(.x$fungicide)),
color = "Experiment") +
theme_minimal() +
theme(
plot.title = element_text(size = 10, hjust = 0.5), # Tamanho do título do gráfico
axis.text.x = element_text(size = 7), # Tamanho do texto do eixo x
axis.text.y = element_text(size = 7), # Tamanho do texto do eixo y
axis.title.x = element_text(size = 7), # Tamanho do título do eixo x
axis.title.y = element_text(size = 7), # Tamanho do título do eixo y
legend.title = element_text(size = 7), # Tamanho do título da legenda
legend.text = element_text(size = 7), # Tamanho do texto da legenda
plot.subtitle = element_text(size = 7), # Tamanho do texto da anotação
# Diminuir o tamanho da fonte da anotação específica para cada gráfico
plot.caption = element_text(size = 5)
)
# Adicionando a anotação de texto no gráfico com o rótulo correspondente ao fungicida
p <- p + annotate("text", x = Inf, y = Inf, hjust = 1, vjust = 1,
label = .y, size = 2.5, color = "black")
# Retornar o gráfico atualizado
return(p)
})
# Use o patchwork para unir os gráficos
final_plot <- wrap_plots(plotlist = plot_list)
# Imprimir o gráfico final
print(final_plot)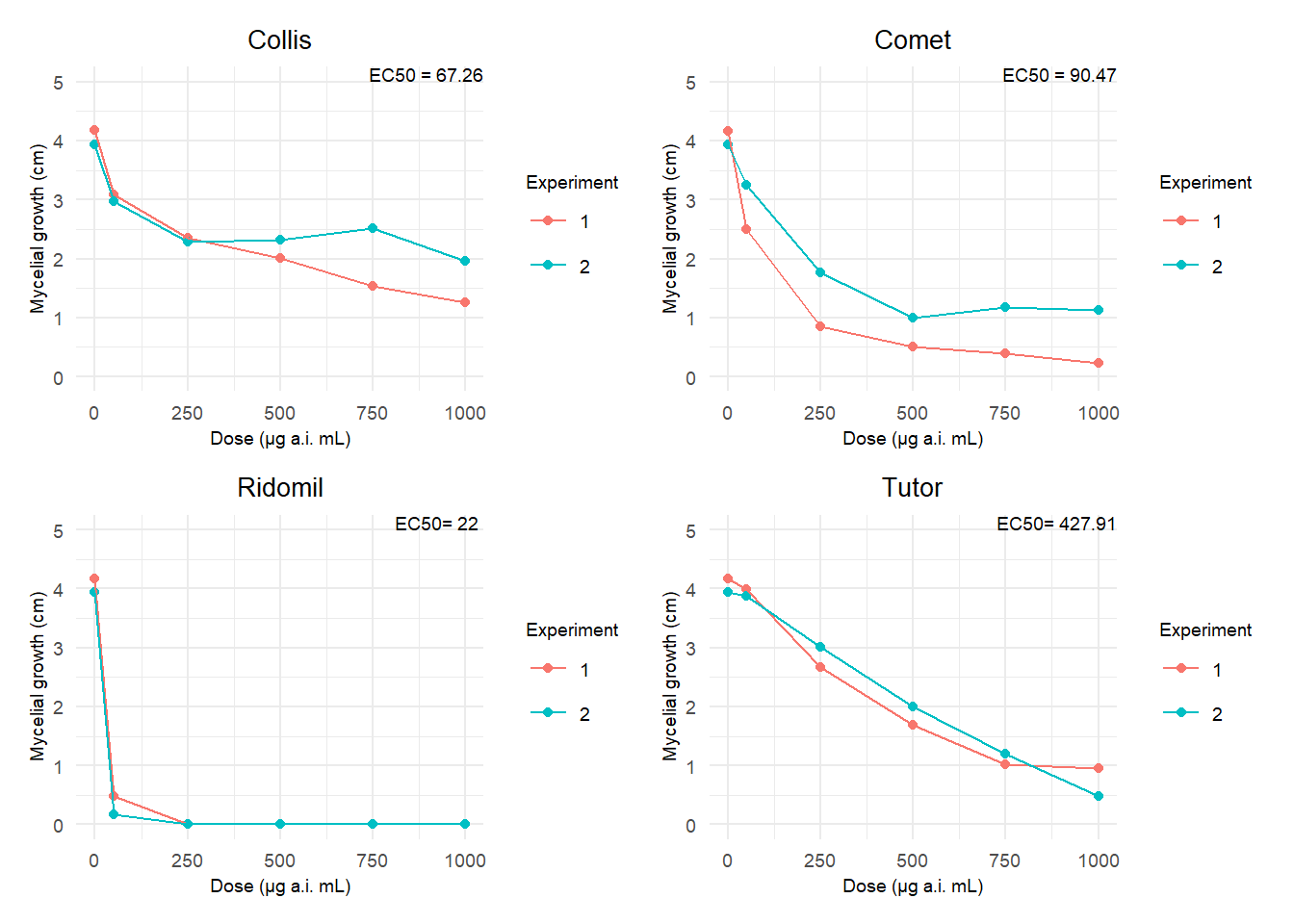
ggsave("plot_vitro.png",
bg = "white", width = 6, height= 4)Model fitting
library(dplyr)
library(drc)
#Ridomil
Ridomil <- fung2 |>
filter(fungicide == "Ridomil")
drcrid <- drm(growth_mean ~ dose, data = Ridomil,
fct = LL.3()) #Dose-Response Analysis
AIC(drcrid) #AIC (Akaike Information Criterion) é uma medida estatística que avalia a qualidade relativa de um modelo estatístico[1] -19.175summary(drcrid)
Model fitted: Log-logistic (ED50 as parameter) with lower limit at 0 (3 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
b:(Intercept) 2.961748 9.264597 0.3197 0.7565
d:(Intercept) 4.052506 0.063675 63.6436 2.945e-13 ***
e:(Intercept) 21.999862 56.542627 0.3891 0.7063
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error:
0.0900501 (9 degrees of freedom)plot(drcrid)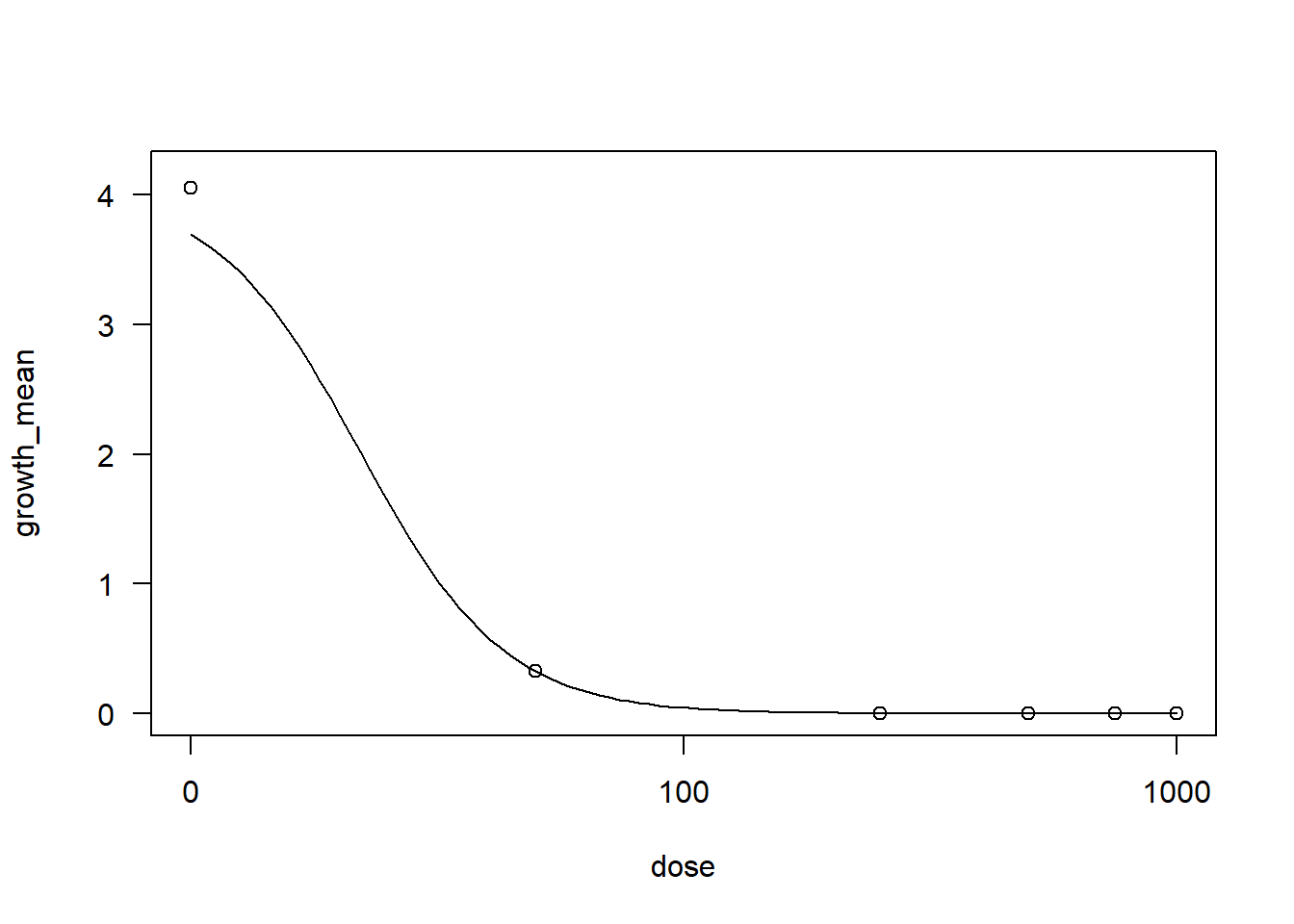
#LL.3() refere-se a uma função log-logística de três parâmetros, comumente usada para modelar respostas a doses crescentes#Collis
Collis <- fung2 |>
filter(fungicide == "Collis")
drccol3 <- drm(growth_mean ~ dose, data = Collis,
fct = LL.3())
AIC(drccol3)[1] 11.25187summary(drccol3)
Model fitted: Log-logistic (ED50 as parameter) with lower limit at 0 (3 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
b:(Intercept) 0.45154 0.11974 3.7711 0.004409 **
d:(Intercept) 4.05505 0.22611 17.9340 2.37e-08 ***
e:(Intercept) 553.36080 195.38485 2.8322 0.019653 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error:
0.319946 (9 degrees of freedom)plot(drccol3)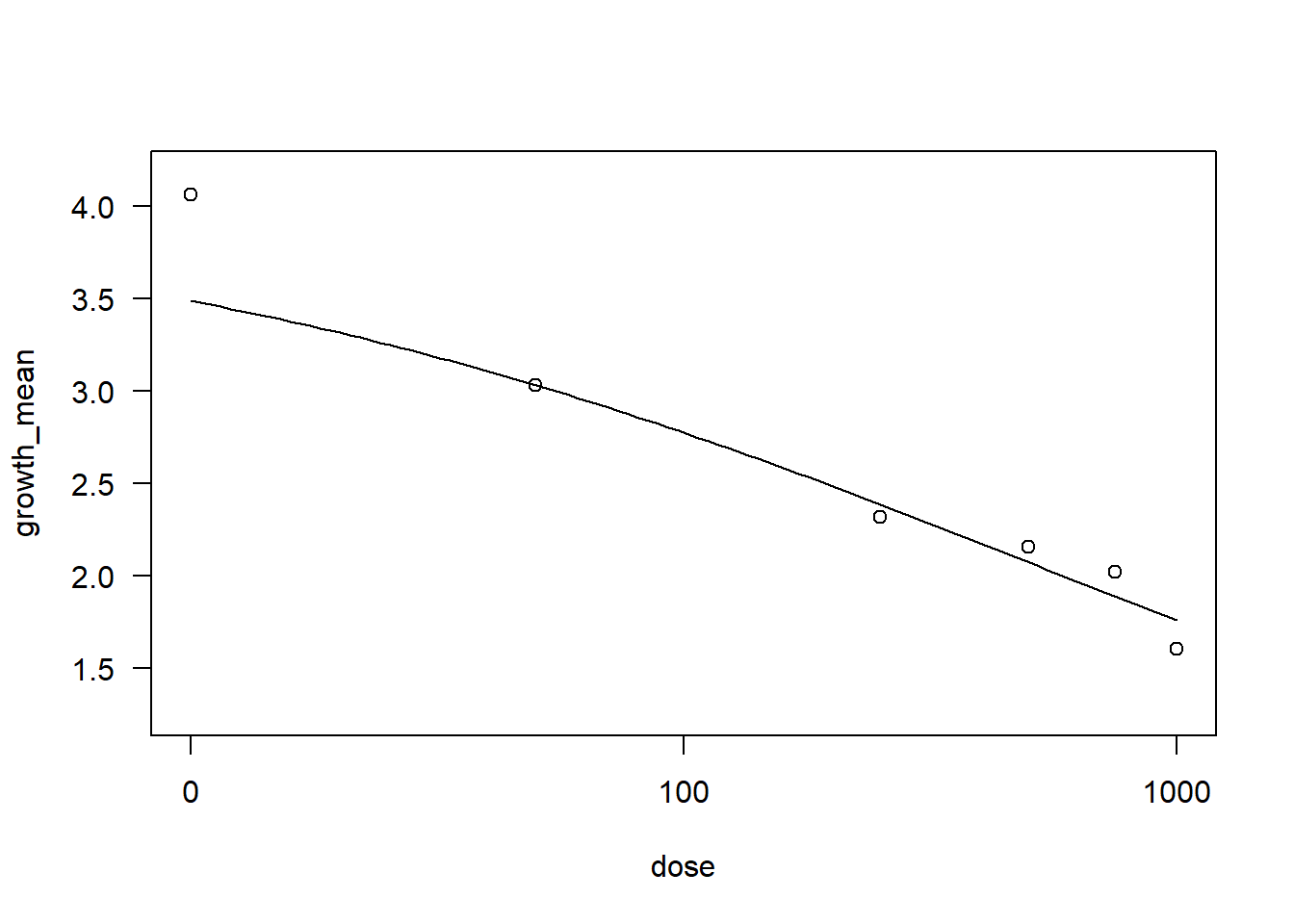
#apenas explorando sem a dose 0 pra ver se ajusta o modelo
Collis2 <- fung2 %>% filter(dose != "0") |>
filter(fungicide == "Collis")
drccol2 <- drm(growth_mean ~ dose, data = Collis2,
fct = LL.3())
AIC(drccol2)[1] 12.18485summary(drccol2)
Model fitted: Log-logistic (ED50 as parameter) with lower limit at 0 (3 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
b:(Intercept) 0.54979 0.68631 0.8011 0.4494
d:(Intercept) 3.64932 2.05307 1.7775 0.1187
e:(Intercept) 844.60241 1645.42755 0.5133 0.6235
Residual standard error:
0.3565222 (7 degrees of freedom)plot(drccol2)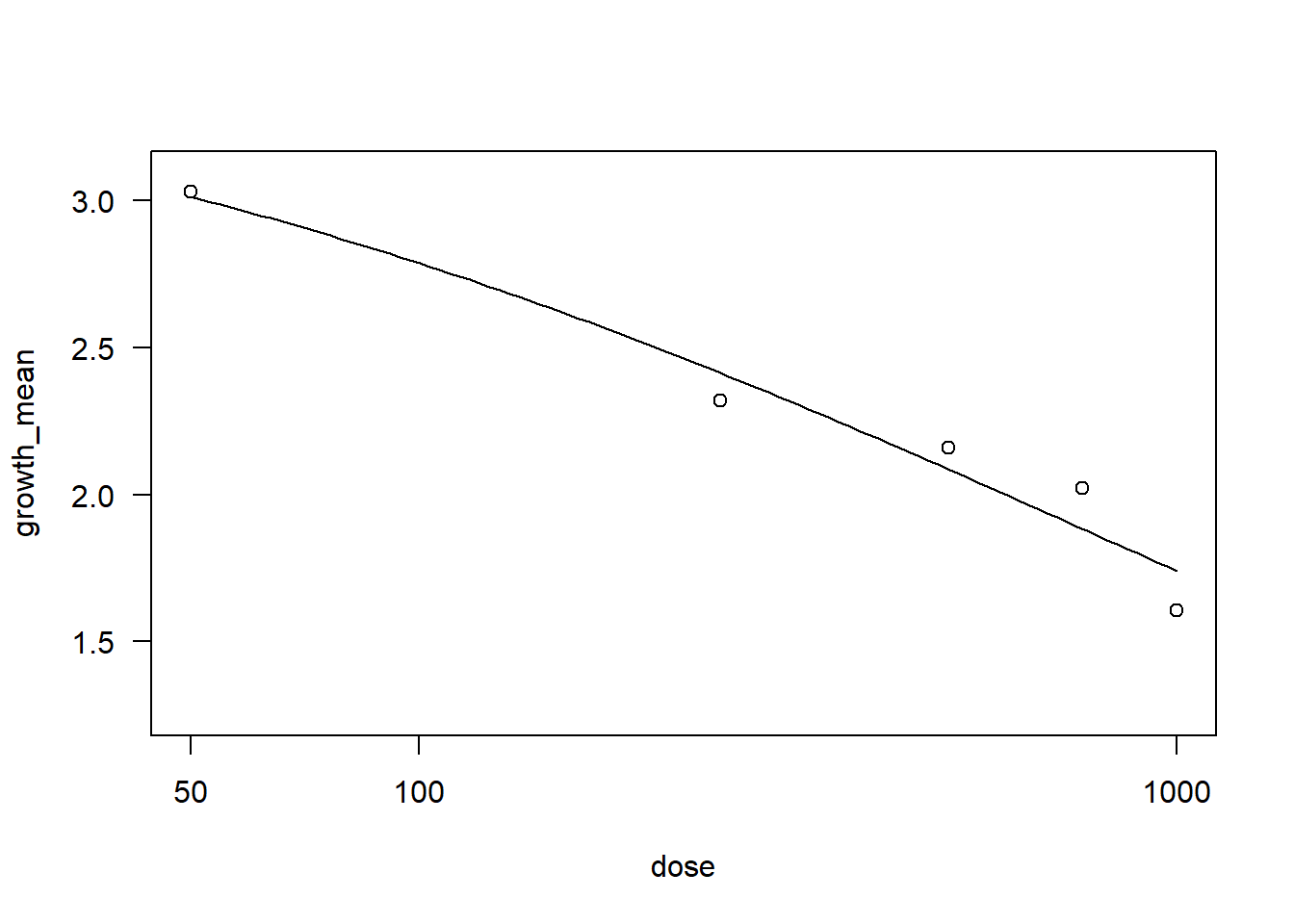
#Model modification
Collis <- fung2 |>
filter(fungicide == "Collis")
drccol <- drm(growth_mean ~ dose, data = Collis,
fct = EXD.3()) #EXP.3:O modelo assume uma relação exponencial entre a resposta e a dose, com três parâmetros estimados para capturar a forma da curva de resposta.
AIC(drccol)[1] 15.16586summary(drccol)
Model fitted: Shifted exponential decay (3 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
c:(Intercept) 1.96620 0.17502 11.2338 1.348e-06 ***
d:(Intercept) 3.98746 0.30126 13.2361 3.326e-07 ***
e:(Intercept) 97.04314 72.46426 1.3392 0.2133
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error:
0.3766195 (9 degrees of freedom)plot(drccol)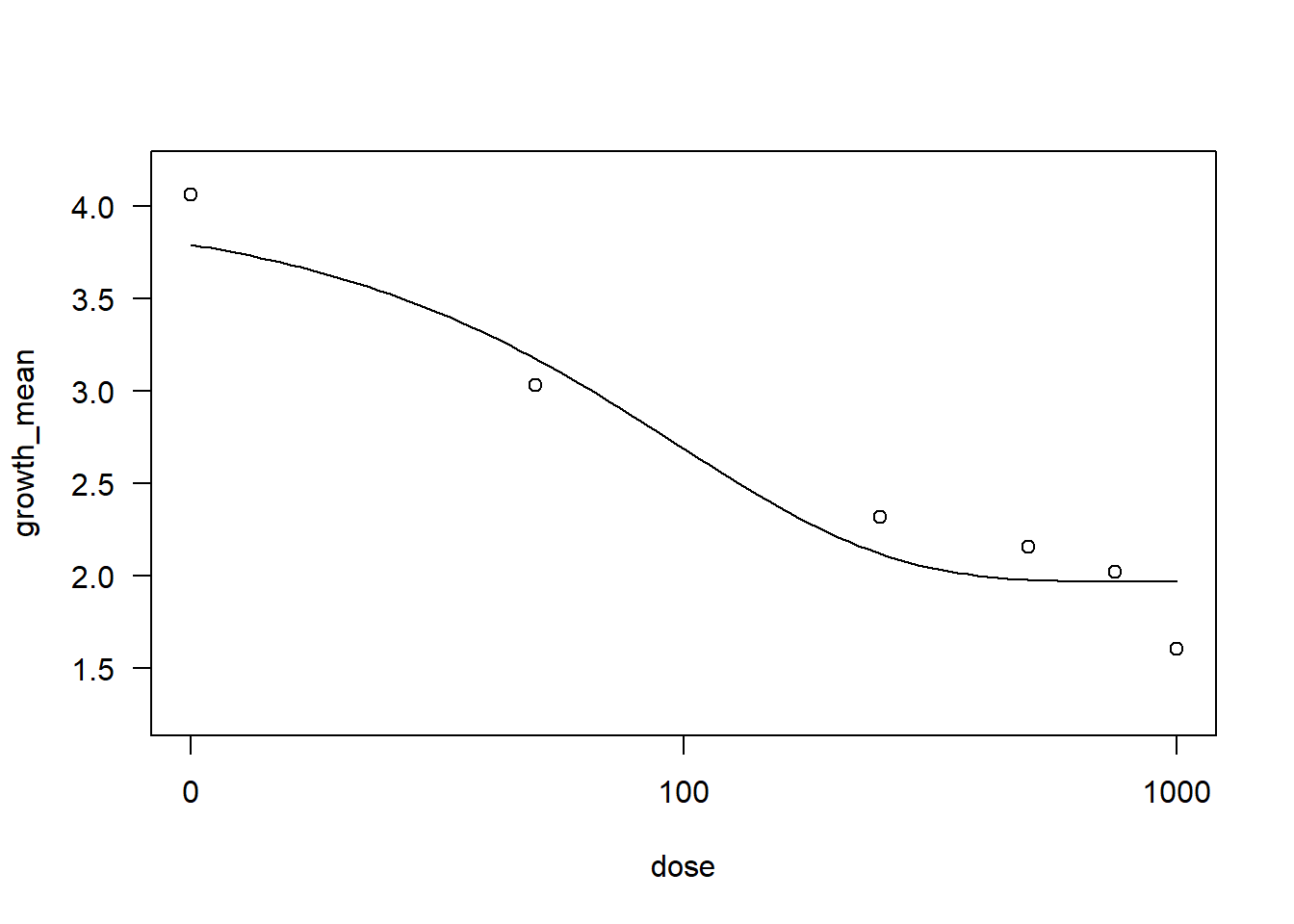
#Tutor
Tutor <- fung2 |>
filter(fungicide == "Tutor")
drctut <- drm(growth_mean ~ dose, data = Tutor,
fct = LL.3())
AIC(drctut)[1] -2.736017summary(drctut)
Model fitted: Log-logistic (ED50 as parameter) with lower limit at 0 (3 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
b:(Intercept) 1.703164 0.170555 9.986 3.620e-06 ***
d:(Intercept) 4.038278 0.099448 40.607 1.659e-11 ***
e:(Intercept) 427.910007 26.487305 16.155 5.913e-08 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error:
0.1786313 (9 degrees of freedom)plot(drctut)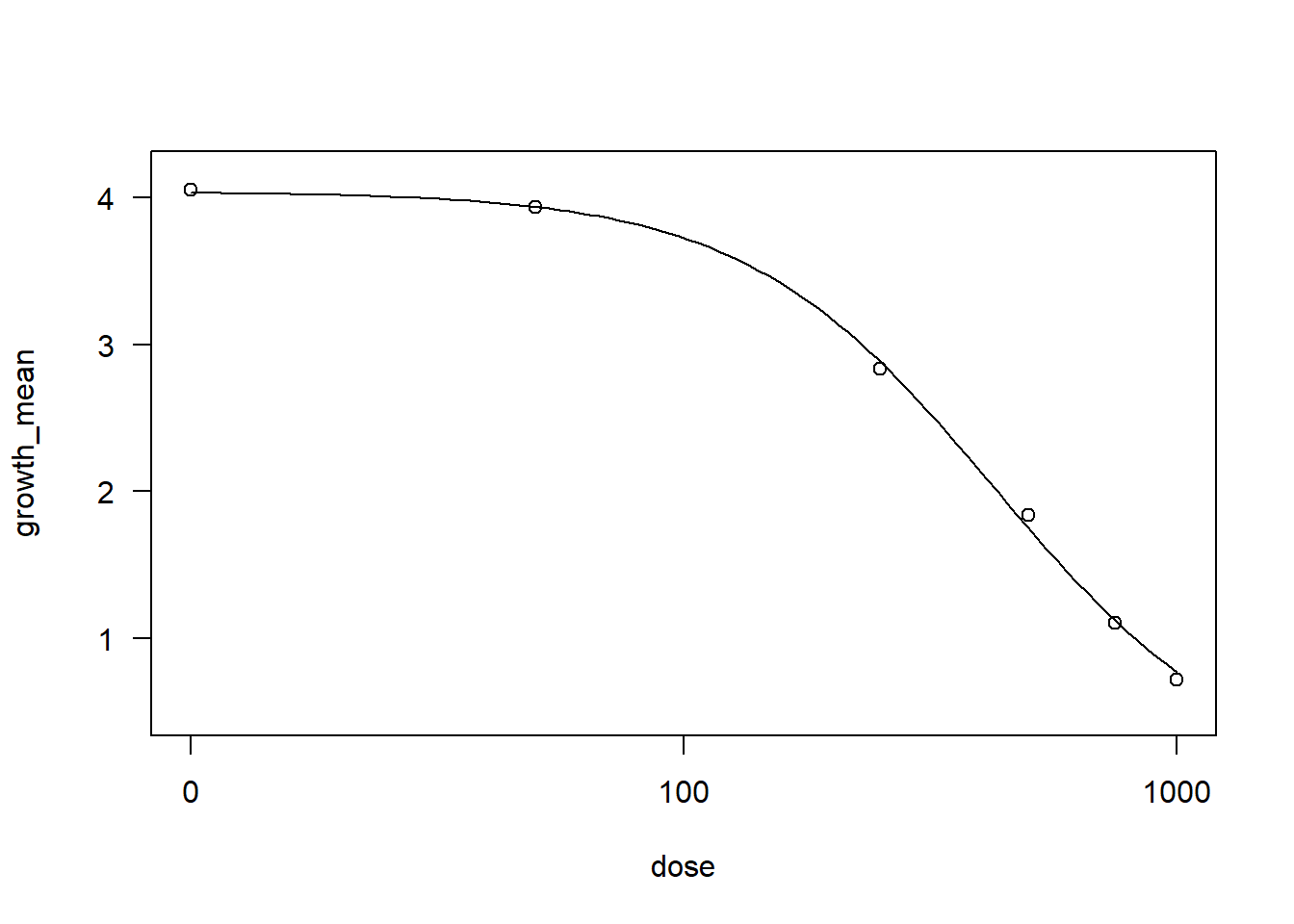
#Comet
Comet <- fung2 |>
filter(fungicide == "Comet")
drccom2 <- drm(growth_mean ~ dose, data = Comet,
fct = LL.3())
AIC(drccom2)[1] 18.56011summary(drccom2)
Model fitted: Log-logistic (ED50 as parameter) with lower limit at 0 (3 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
b:(Intercept) 0.89510 0.18828 4.7541 0.001038 **
d:(Intercept) 4.07136 0.30265 13.4525 2.893e-07 ***
e:(Intercept) 121.70989 37.58603 3.2382 0.010188 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error:
0.4338342 (9 degrees of freedom)plot(drccom2)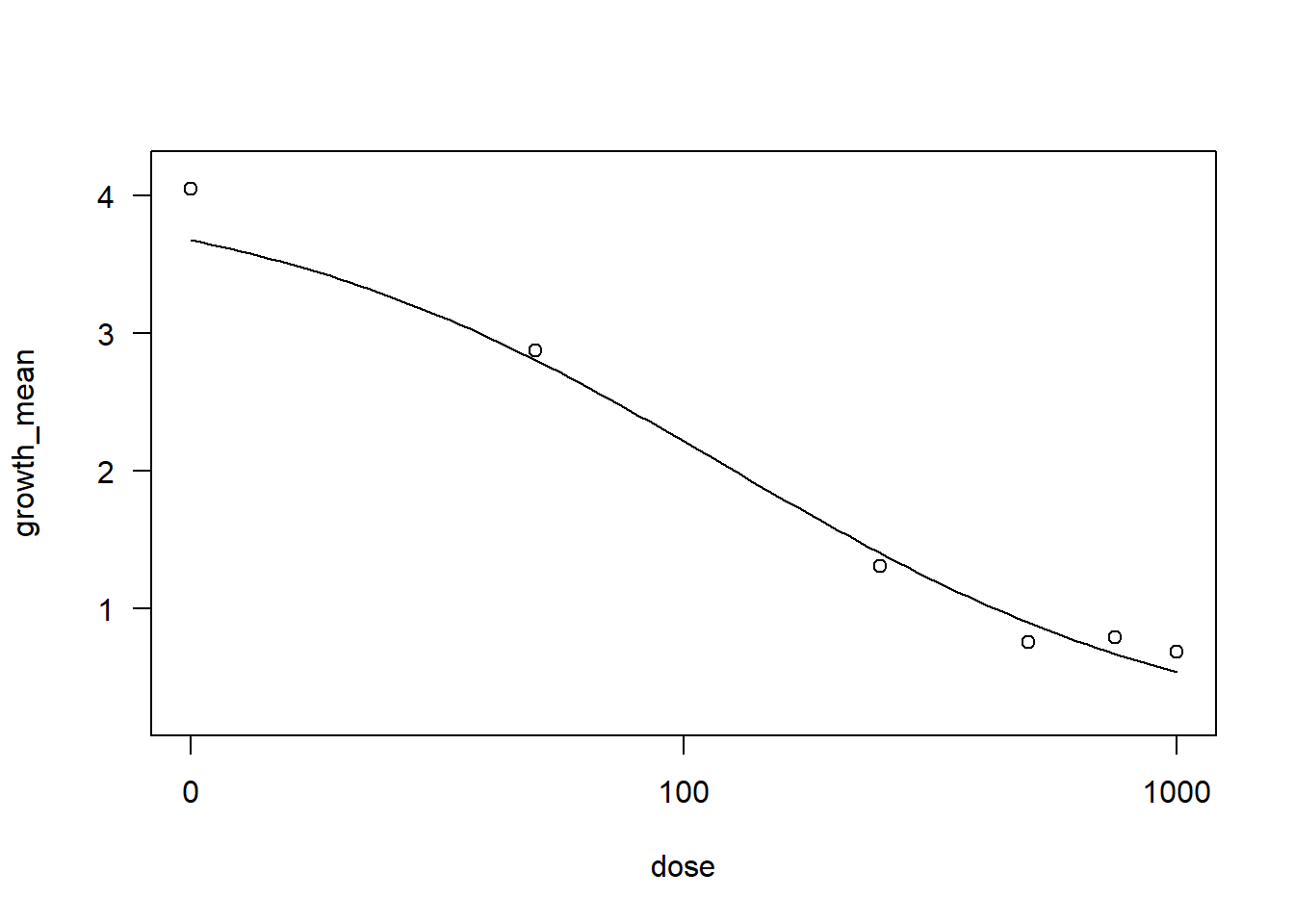
drccom <- drm(growth_mean ~ dose, data = Comet,
fct = EXD.3())
AIC(drccom)[1] 17.92899summary(drccom)
Model fitted: Shifted exponential decay (3 parms)
Parameter estimates:
Estimate Std. Error t-value p-value
c:(Intercept) 0.72856 0.19049 3.8247 0.004061 **
d:(Intercept) 4.00455 0.28239 14.1807 1.836e-07 ***
e:(Intercept) 130.52175 43.37518 3.0091 0.014737 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error:
0.4225745 (9 degrees of freedom)plot(drccom)
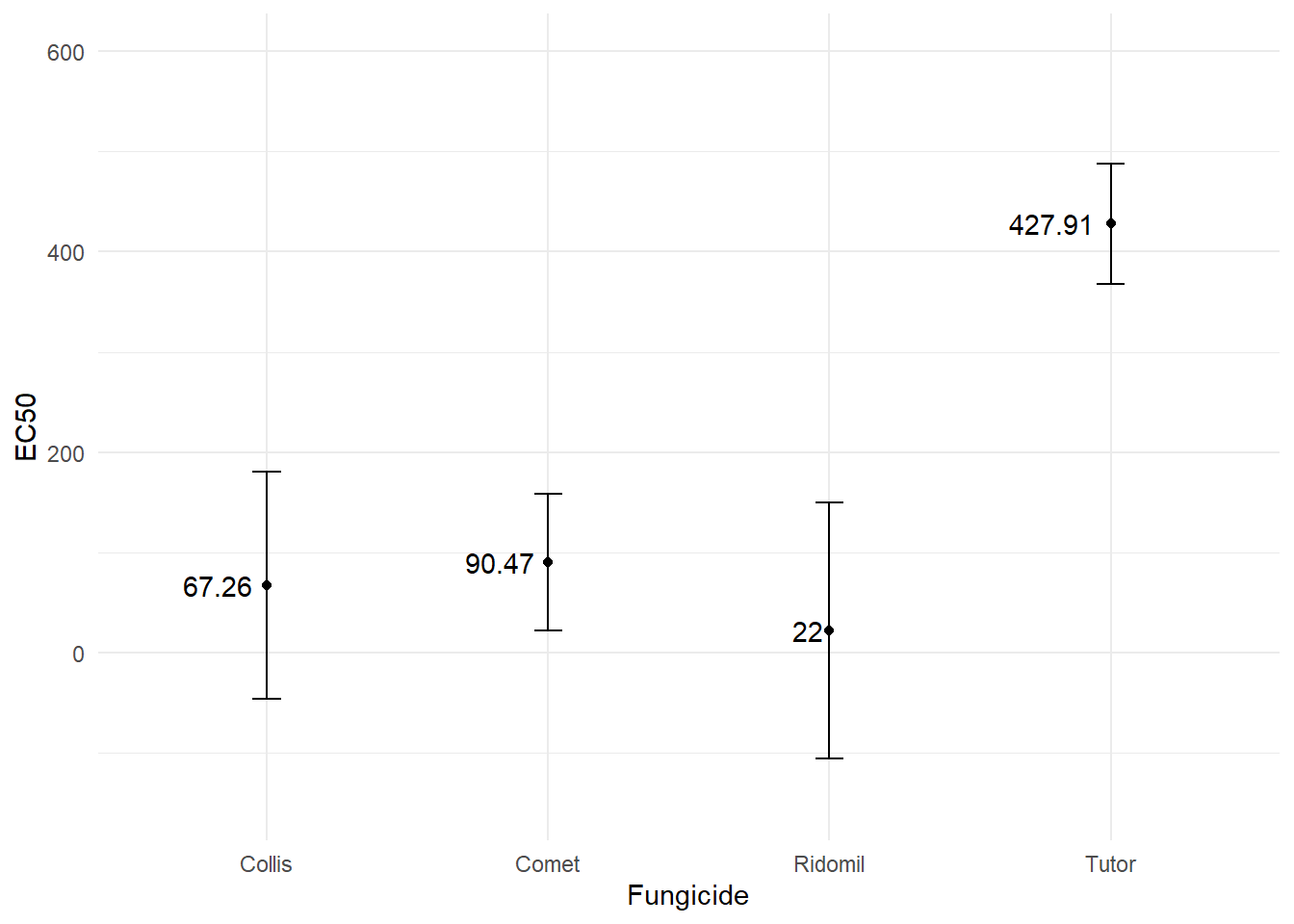
EC50
erid <- ED(drcrid, 50, interval = "delta") #ridomil
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 22.000 56.543 -105.908 149.908ecol <- ED(drccol, 50, interval = "delta") #collis
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 67.265 50.228 -46.359 180.890etut <- ED(drctut, 50, interval = "delta") #tutor
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 427.910 26.487 367.992 487.828ecom <- ED(drccom, 50, interval = "delta") #comet
Estimated effective doses
Estimate Std. Error Lower Upper
e:1:50 90.471 30.065 22.458 158.483dadosec <- data.frame( Trat = c("Ridomil", "Collis", "Tutor", "Comet"),
Estimate = c(22.00, 67.265, 427.910, 90.471),
Lower = c(-105.908, -46.359, 367.992, 22.458),
Upper = c(149.908, 180.890, 487.828, 158.483))
datatable(dadosec)library(ggplot2)
plotEC <- dadosec |>
ggplot(aes(Trat, Estimate)) +
geom_point() +
ylim(-150,600)+
geom_errorbar(aes(ymin = Lower, ymax = Upper), width = 0.1) +
geom_text(aes(label = round(Estimate, 2)), hjust = 1.2) +
labs(x = "Fungicide", y = "EC50") +
theme_minimal()
plotEC